New Technique Gives Researchers a Near-atom-level View of Enzyme’s Role in Correcting Misfolded Proteins
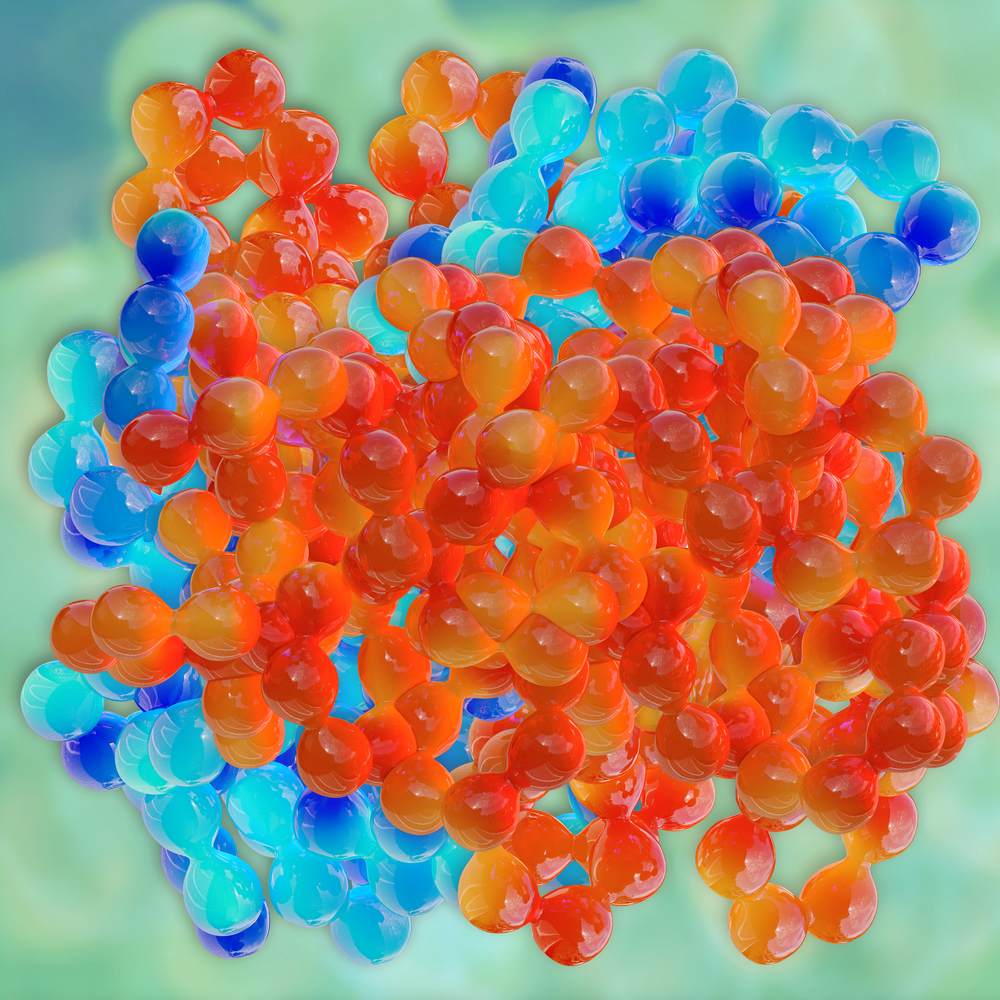
University of Michigan and University of Pennsylvania researchers used a cutting-edge technique called cryo-electron microscopy to see how the Hsp104 yeast enzyme breaks up clumps of the misfolded proteins that are hallmarks of neurodegenerative diseases like ALS.
The resolution that the technique provided was so high that it was close to the level of individual atoms.
Researchers discovered that Hsp104 pulls misfolded proteins in through a central channel to process them in a way that leads to the proteins refolding in the proper manner. Proteins contain components called polypeptides, and Hsp104 retrieves one polypeptide at a time from a clump of misfolded polypeptides. Once Hsp104 releases the polypeptide, it is free to refold in the proper manner, or the cell machinery can degrade it.
The study, “Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104,” was published in the journal Science.
In amyotrophic lateral sclerosis, Alzheimer’s, and other neurodegenerative disorders, the main disease-triggering event is the formation of misfolded proteins. Those proteins tend to gravitate to normal proteins, causing them to misfold as well. The chain of events propagates the disease.
If scientists could find a way to unravel misfolded proteins, they could stop the disease. Until the Michigan and Pennsylvania research, they had not learned how to do it.
Scientists have dubbed Hsp104 a disaggregase enzyme because studies have shown it can de-clump aggregated protein, helping them obtain the correct shape.
Interestingly, Hsp104 is found in most of the Earth’s organisms except animals. But researchers theorized that a drug centered around Hsp104 could help pull apart misfolded proteins in humans, helping to heal neurodegenerative diseases.
Dr. James Shorter’s lab at the University of Pennsylvania had previously established that a version of Hsp104 can rectify the misfolded proteins involved in neurodegenerative diseases.
Taking the work a step further, Shorter’s lab and Dr. Daniel R. Southworth’s lab at the University of Michigan collaborated on a way to obtain a high-resolution view of Hsp104’s structure.
Shorter’s lab created the highly purified Hsp104 enzymes used in the research. Southworth’s lab used cryo-electron microscopy to deliver the clearest image of Hsp104 to date. It showed how the enzyme de-clumps protein aggregates.
“This superb collaboration has yielded the highest resolution picture of Hsp104 caught in the act of processing a protein,” Shorter, an associate professor of biochemistry and biophysics at the University of Pennsylvania’s Perelman School of Medicine, said in a press release. “We can now see the moving parts of the Hsp104 complex and how we might tune it to optimally attack neurodegenerative disease proteins.”
Hsp104 pulled the polypeptides through “stepwise, like a ratchet,” said Southworth, an assistant professor at the University of Michigan’s Life Sciences Institute. “We can see how the proteins in the machine rearrange between different states to grab onto the next site on the substrate,” or substance upon which an enzyme acts, he said.
Being able to observe the enzyme’s action in such a highly magnified way has given the researchers an idea of which parts of the un-clumping mechanism they might manipulate to produce neurodegenerative-disease therapies.
In fact, they have already engineered changes in Hsp104 to enhance its activity toward the proteins involved in neurodegenerative diseases.
“The study helps us to understand how cells can break apart toxic protein aggregates to restore protein function,” Shorter said. “Finally having a clear picture of this remarkable nanomachine will empower our designs for therapeutic versions that work in humans.”